 |
 |
Research Project P9:
Tracking Stem Cell Genealogical tree in Microwells
Research Project Goal
The goal of this project is to use time-lapse microscopy to trackdivisional history of single stem cells over time. Individual cells are confined within an array of single microwells and over time the cell divides multiple times to yield a population of cells within each microwell. In this project, we want to use time-lapse microscopy images to determine the divisional pattern of single stem cells, meaning the number of times each cell has divided and which daughter cells came from the division.
Adult stem cells are the cells responsible for growth, repair and regeneration of tissues after birth and they are found in many different tissues, including brain, muscle, pancreas, skin, etc. These cells have great therapeutic potential because they can both self-renew, to make more stem cells, and differentiate, to form all the specialized cells of the tissue. One example of this therapeutic use is bone marrow transplants. Bone marrow contains hematopoietic stem cells, which can regenerate all the blood cell lineages. However, the use of stem cells for therapy is limited because there are very few stem cells in tissue. If the cells are plated in culture in hopes that they will divide and form more stem cells, the stem cells tend to lose their "stemmess," so they are no longer capable of regeneration.
In the Blau lab, we are working to create an environment, or artificial niche, that will enable the expansion of stem cells, such that when cells are plated in culture and divide, they will maintain their "stemmess." Towards this goal, we are studying single cells in microwells in order to understand division kinetics at a more fundamental level and testing the impact that different protein signaling has the kinetics of division. In previous work, we have manually counted the number of cells in thousands of different wells. This process is very time consuming! In addition these movies contain a HUGE amount of data and we are currently only utilizing a tiny portion of their potential. We are hoping to automate the cell counting process and proceed to more advanced analysis of this enormous volume of data.
Research Project Scope
Given time-lapse video of stem cells in microwells, track cell movement and division over time.
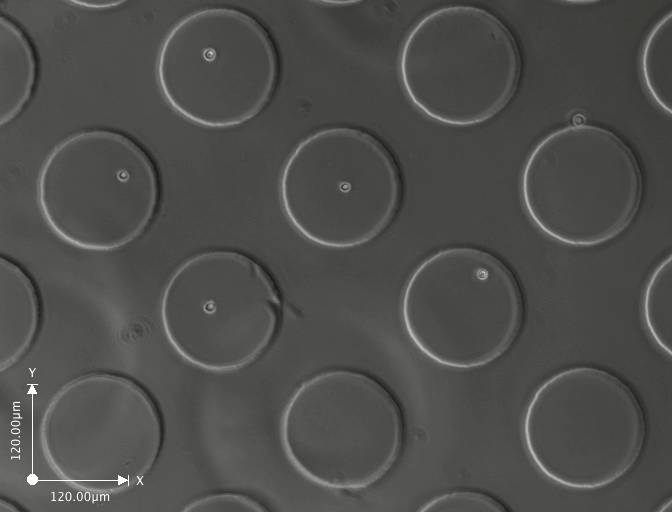 Day 1 (click on image to enlarge)
Day 1 (click on image to enlarge)
 Day 2 (click on image to enlarge)
Day 2 (click on image to enlarge)
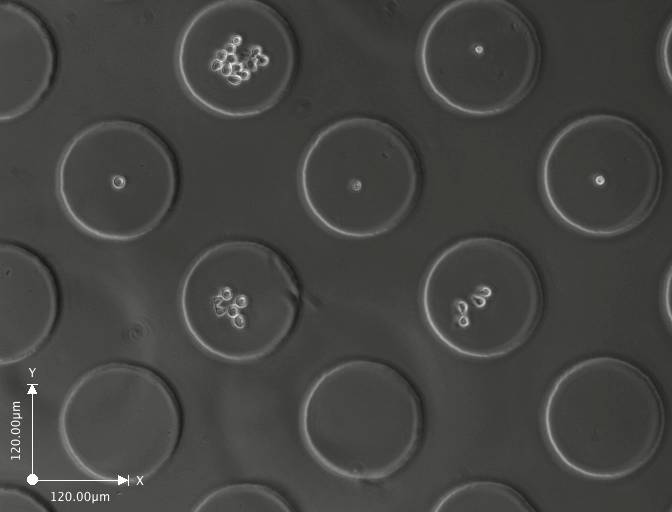 Day 3 (click on image to enlarge)
Day 3 (click on image to enlarge)
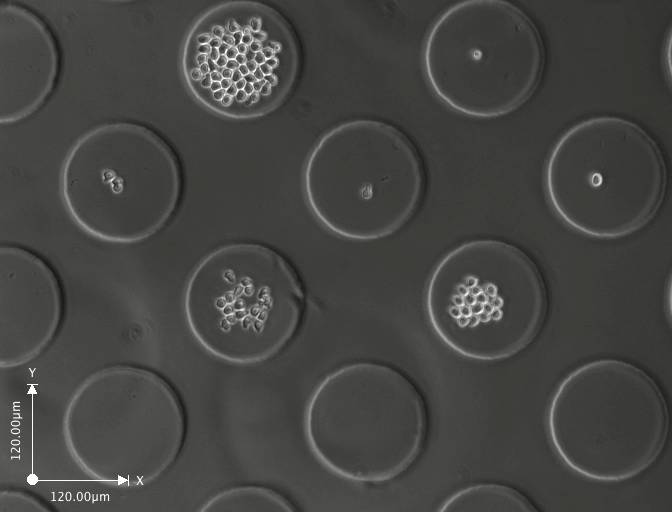 Day 4 (click on image to enlarge)
Day 4 (click on image to enlarge)
These are a few images from a time-lapse movie showing the cells dividing and increasing in number. The movie consists of images taken at regular intervals for 3-5 days.
Tasks
Track divisional history of single stem cells
- Automatically annotate cells
- Give divisional history (genealogical tree)
- Evaluate cell death
- Organize data such that it can be searched based on parameters and used for statistical analysis
Research Project Status
student names here
Point of Contact
Karen Havenstrite: khaven@stanford.edu
Midterm Report
not yet submited
Final Report
not yet submitted
|
 |
|
|

