Research Project P15:
Robotic Image Mosaicing for Optical Biopsy
Research Project Goal
In the Clark Center Biorobotics Lab we are working on an optical biopsy system using a micro-endoscope attached to a Phantom robot for building image mosaics. The Phantom provides information about image overlap as well as the probes position and orientation. We are collaborating with physicians and plan to apply this technology in the clinic.
One of the main challenges of this project is dealing with sensor error, which can occur due to patient/tissue motion and can accumulate over a long exploration path. This problem is analogous to SLAM (simultaneous localization and mapping), the problem of using a mobile robot to estimate both a map of its environment and its pose using noisy sensor data. The goal of this project is to draw on techniques from SLAM and GraphSLAM to deal with sensor noise in the optical biopsy system.
While the main goal of this project is to make the 2D mosaicing system more robust, there may also be opportunities for working with 3D data sets for volume mosaicing.
Research Project Scope
We have already developed algorithms for mosaicing the images in real time at a rate of roughly 1-2 images per second. By extending these current algorithms to handle sensor error, we will make the system more robust and applicable for clinical procedures.
Sample Data
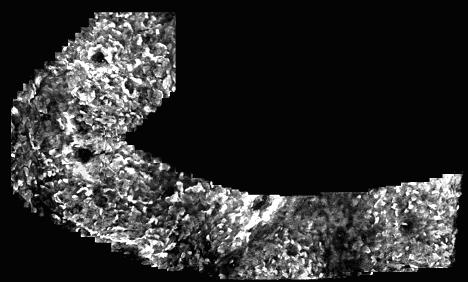
Image mosaic of a human hand, acquired using a Mauna Kea CellVizio micro-endoscope. We already have several sets of data similar to this, consisting of video sequences from the CellVizio, Phantom sensor data, and timestamps for synchronization.
Tasks
- Develop a SLAM model for the optical biopsy system
- Incorporate camera images and Phantom data into the model
- Test the model on data sets where the current algorithms fail
- Apply and adapt 2-D techniques to volumetric data for 3-D mosaics
Research Project Status
Kevin Loewke
Point of Contact
Sebastian Thrun and Kevin Loewke (kloewke@stanford.edu)
Midterm Report
not yet submitted
Final Report
not yet submitted
|

